Real-time reporting of vaccine target recommendations for the COVID-19 coronavirus (SARS-CoV-2)
COVIDep provides an up-to-date set of B-cell and T-cell epitopes that can serve as potential vaccine targets for Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2), the virus causing the COVID-19 pandemic. The identified epitopes are experimentally-derived from SARS-CoV (the virus that caused the 2003 SARS outbreak) and have a close genetic match with the available SARS-CoV-2 sequences.
We thank all the authors, the originating and submitting laboratories for sharing SARS-CoV-2 genetic sequences and SARS-CoV epitopes through the GISAID and VIPR databases, respectively. We also thank the IEDB database for providing the population coverage estimation tool for T cell epitopes.
Related publication: Ahmed, S.F., Quadeer, A.A. & McKay, M.R. COVIDep: a web-based platform for real-time reporting of vaccine target recommendations for SARS-CoV-2. Nature Protocols (2020).
Epitopes identified as potential SARS-CoV-2 vaccine targets
© 2020 The Hong Kong University of Science and Technology. All rights reserved.
Fraction of sequences with mutations in the S protein
Reference sequence of the S protein
Mutations at each position of the S protein
Fraction of sequences with mutations in the E protein
Reference sequence of the E protein
Mutations at each position of the E protein
Fraction of sequences with mutations in the M protein
Reference sequence of the M protein
Mutations at each position of the M protein
Fraction of sequences with mutations in the N protein
Reference sequence of the N protein
Mutations at each position of the N protein
Fraction of sequences with mutations in the orf1a protein
Reference sequence of the orf1a protein
Mutations at each position of the orf1a protein
Fraction of sequences with mutations in the orf1b protein
Reference sequence of the orf1b protein
Mutations at each position of the orf1b protein
Fraction of sequences with mutations in the ORF3a protein
Reference sequence of the ORF3a protein
Mutations at each position of the ORF3a protein
Fraction of sequences with mutations in the ORF6 protein
Reference sequence of the ORF6 protein
Mutations at each position of the ORF6 protein
Fraction of sequences with mutations in the ORF7a protein
Reference sequence of the ORF7a protein
Mutations at each position of the ORF7a protein
Fraction of sequences with mutations in the ORF7b protein
Reference sequence of the ORF7b protein
Mutations at each position of the ORF7b protein
Fraction of sequences with mutations in the ORF8 protein
Reference sequence of the ORF8 protein
Mutations at each position of the ORF8 protein
Fraction of sequences with mutations in the ORF10 protein
Reference sequence of the ORF10 protein
Mutations at each position of the ORF10 protein
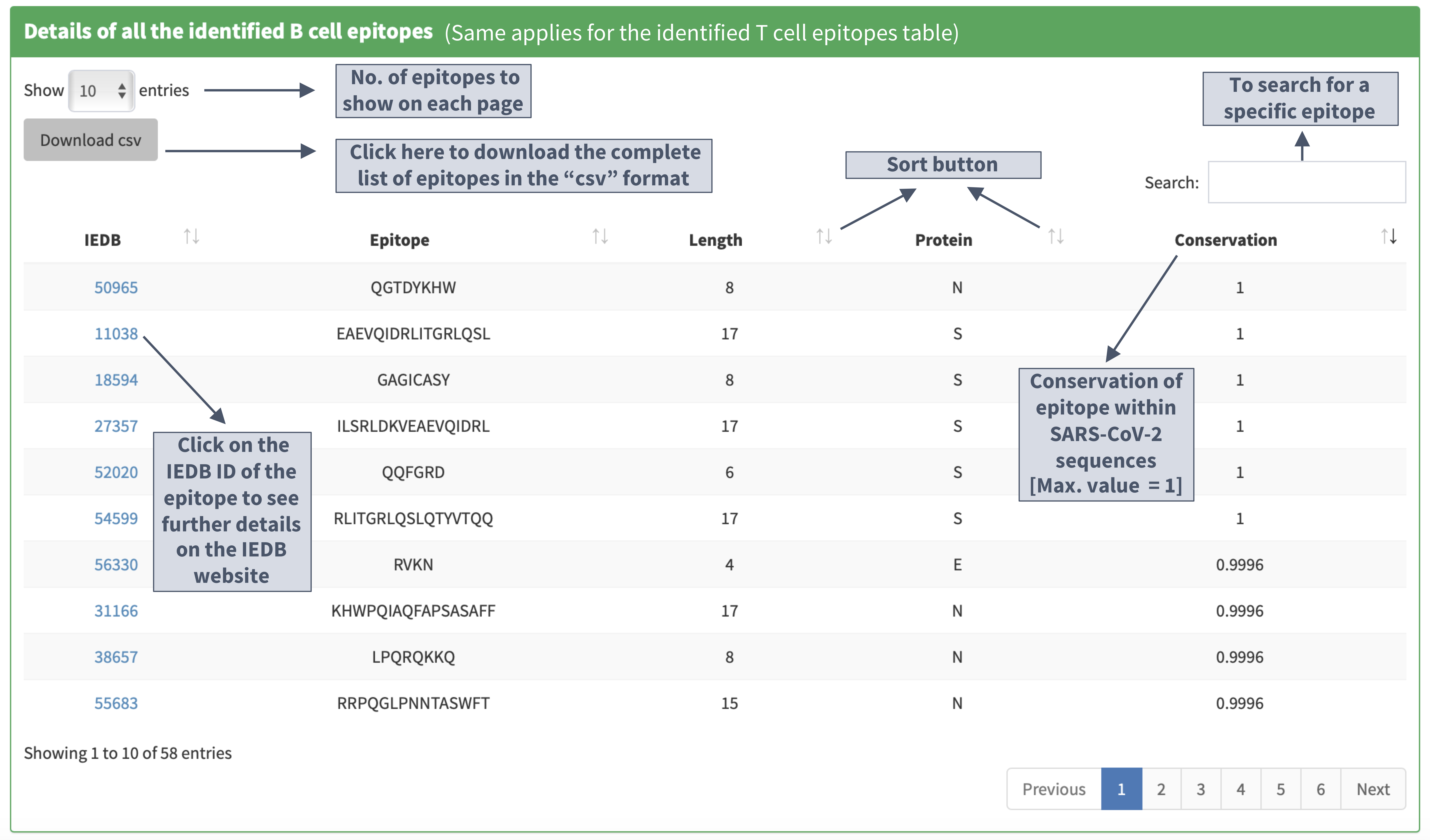
Details of all the identified B cell epitopes
Location of the identified B cell epitopes on the primary structure of the S protein
Details of the identified B cell epitopes in the S protein
Location of the identified B cell epitopes on the primary structure of the E protein
Details of the identified B cell epitopes in the E protein
Location of the identified B cell epitopes on the primary structure of the M protein
Details of the identified B cell epitopes in the M protein
Location of the identified B cell epitopes on the primary structure of the N protein
Details of the identified B cell epitopes in the N protein
Location of the identified B cell epitopes on the primary structure of the orf1a protein
Details of the identified B cell epitopes in the orf1a protein
Location of the identified B cell epitopes on the primary structure of the orf1b protein
Details of the identified B cell epitopes in the orf1b protein
Location of the identified B cell epitopes on the primary structure of the ORF3a protein
Details of the identified B cell epitopes in the ORF3a protein
Location of the identified B cell epitopes on the primary structure of the ORF6 protein
Details of the identified B cell epitopes in the ORF6 protein
Location of the identified B cell epitopes on the primary structure of the ORF7a protein
Details of the identified B cell epitopes in the ORF7a protein
Location of the identified B cell epitopes on the primary structure of the ORF7b protein
Details of the identified B cell epitopes in the ORF7a protein
Location of the identified B cell epitopes on the primary structure of the ORF8 protein
Details of the identified B cell epitopes in the ORF8 protein
Location of the identified B cell epitopes on the primary structure of the ORF10 protein
Details of the identified B cell epitopes in the ORF10 protein
0
Identified discontinuous B cell epitopes
Details of all the identified T cell epitopes
Location of the identified T cell epitopes on the primary structure of the S protein
Details of the identified T cell epitopes in the S protein
Location of the identified T cell epitopes on the primary structure of the E protein
Details of the identified T cell epitopes in the E protein
Location of the identified T cell epitopes on the primary structure of the M protein
Details of the identified T cell epitopes in the M protein
Location of the identified T cell epitopes on the primary structure of the N protein
Details of the identified T cell epitopes in the N protein
Location of the identified T cell epitopes on the primary structure of the orf1a protein
Details of the identified T cell epitopes in the orf1a protein
Location of the identified T cell epitopes on the primary structure of the orf1b protein
Details of the identified T cell epitopes in the orf1b protein
Location of the identified T cell epitopes on the primary structure of the ORF3a protein
Details of the identified T cell epitopes in the ORF3a protein
Location of the identified T cell epitopes on the primary structure of the ORF6 protein
Details of the identified T cell epitopes in the ORF6 protein
Location of the identified T cell epitopes on the primary structure of the ORF7a protein
Details of the identified T cell epitopes in the ORF7a protein
Location of the identified T cell epitopes on the primary structure of the ORF7b protein
Details of the identified T cell epitopes in the ORF7b protein
Location of the identified T cell epitopes on the primary structure of the ORF8 protein
Details of the identified T cell epitopes in the ORF8 protein
Location of the identified T cell epitopes on the primary structure of the ORF10 protein
Details of the identified T cell epitopes in the ORF10 protein
Region
MHC allele class
Assay type
COVIDep is made possible by the open sharing of genome sequence data of SARS-CoV-2 sequences by research groups from around the world through the GISAID platform, and the open sharing of immunological data of experimentally determined SARS-CoV epitopes through IEDB and ViPR databases. We gratefully acknowledge the contributions of all the researchers, scientists and technical staff involved.
We would like to acknowledge the support extended by the Department of Electronic and Computer Engineering at the Hong Kong University of Science and Technology for providing the required resources and setting up the server for the web application. We especially thank Nelvin Law and Kenny Pang for their technical support.
Special thanks to Raymond Louie, David Morales, Saqib Sohail, Neelkanth Kundu, Awais Shah, and Umer Abdullah for comments and suggestions that helped to improve the web application and its interface.
Acknowledgement for all epitope data downloaded from ViPR/IEDB
Acknowledgement for all the R/Shiny tools and packages used in the development
Home
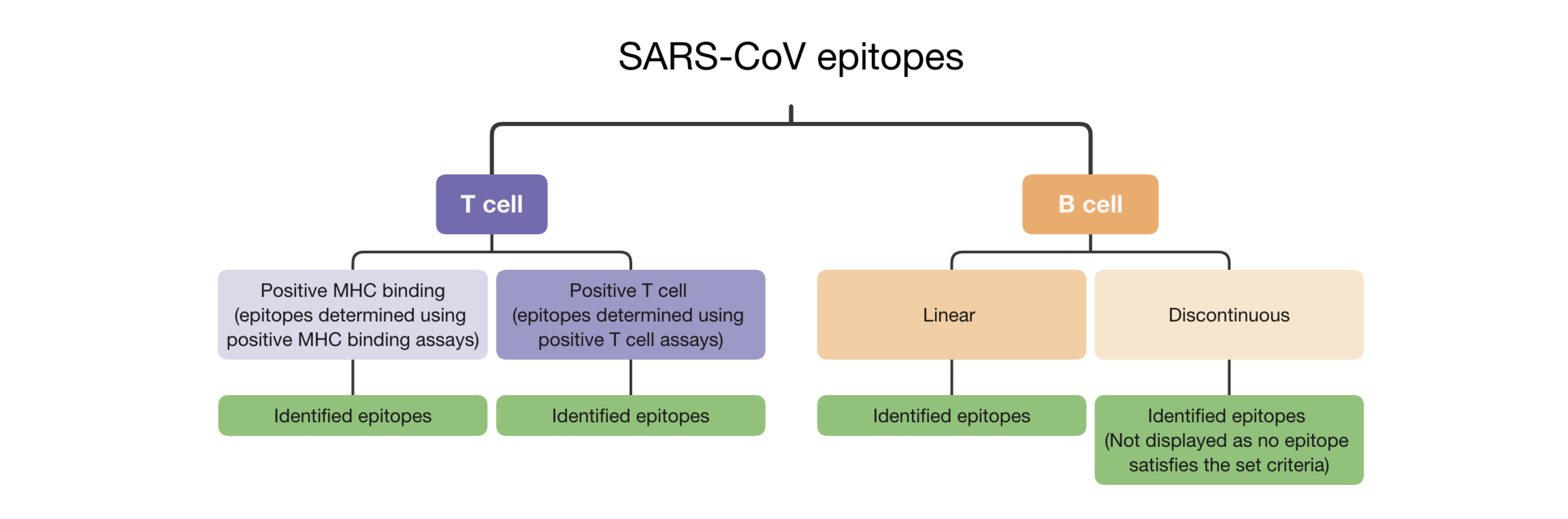
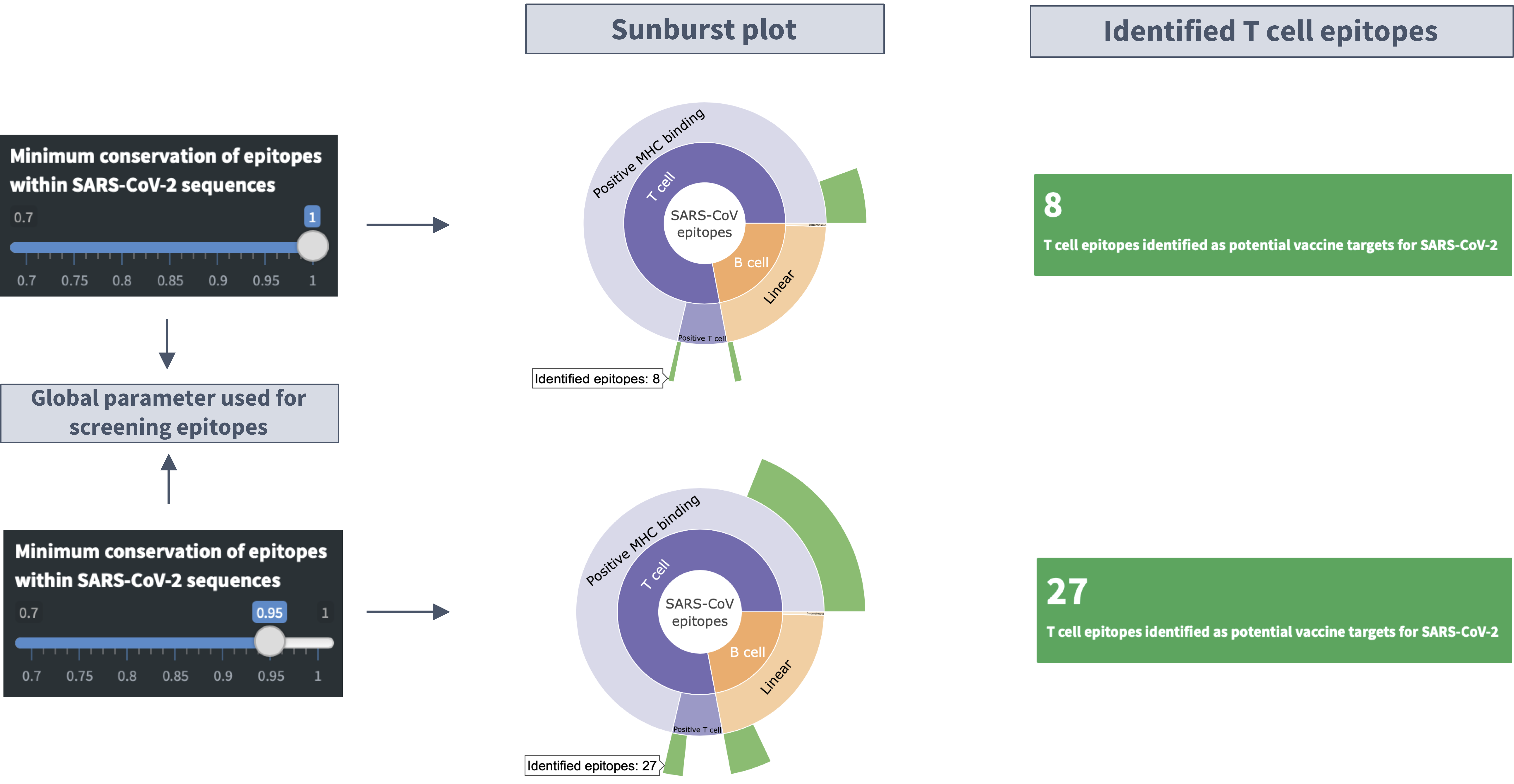
Description of the 'sunburst' plot
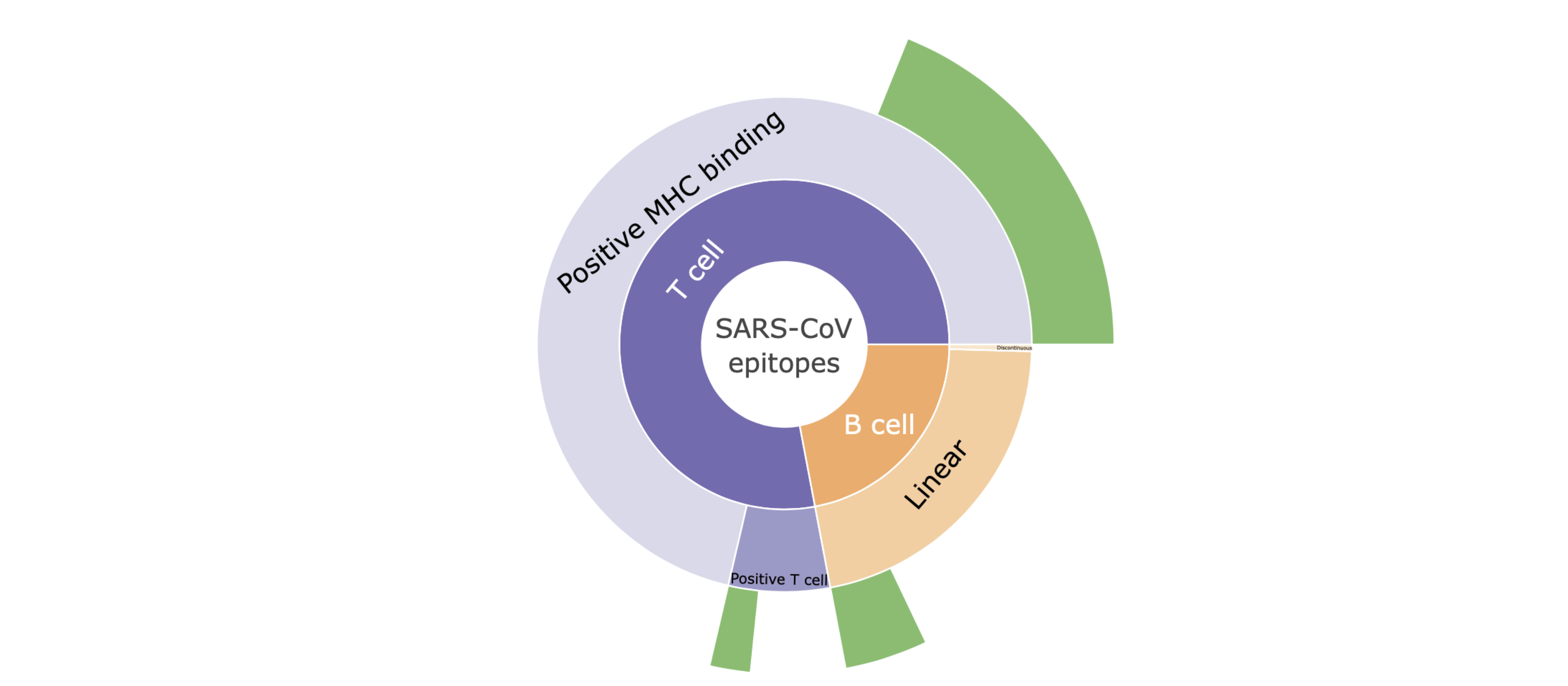
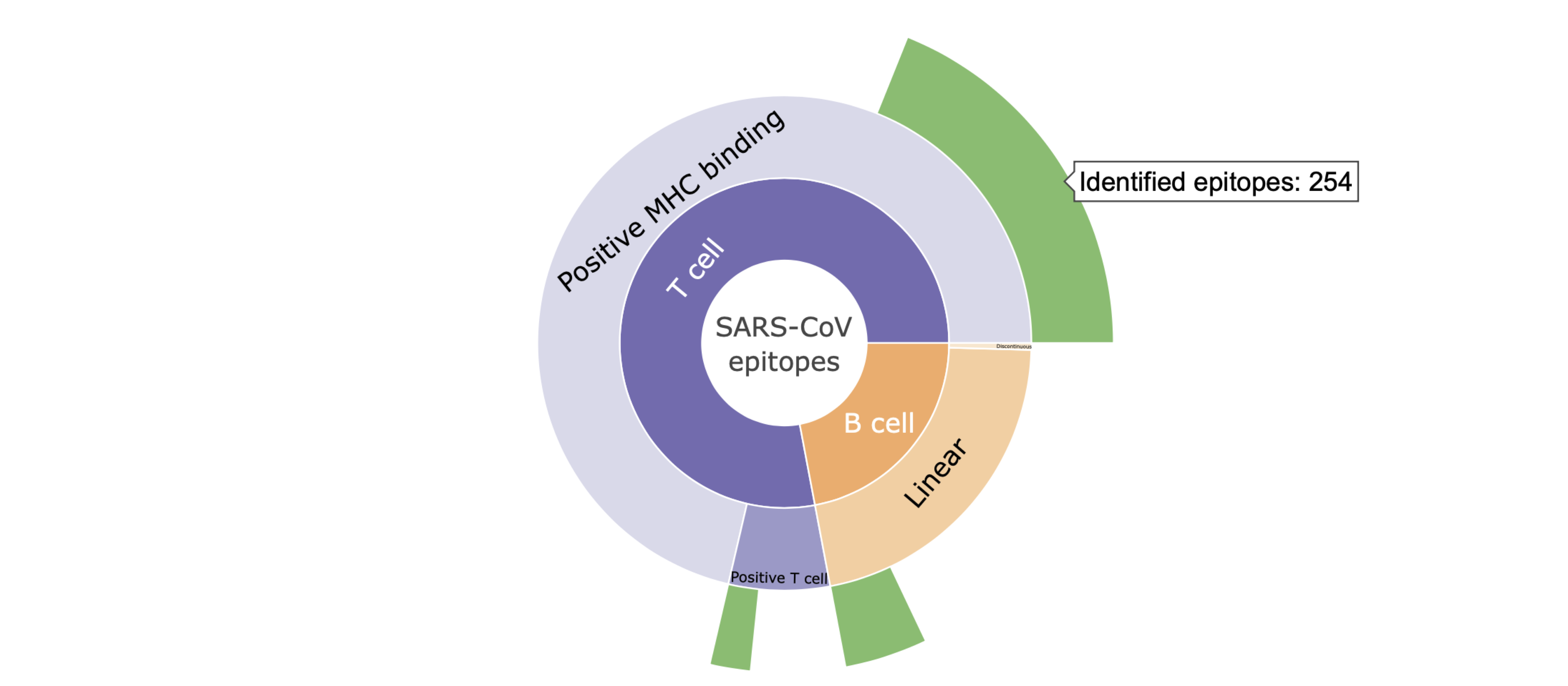
I. Hovering mouse over the plot shows the specific number of the SARS-CoV epitopes with high genetic match in SARS-CoV-2 sequences.
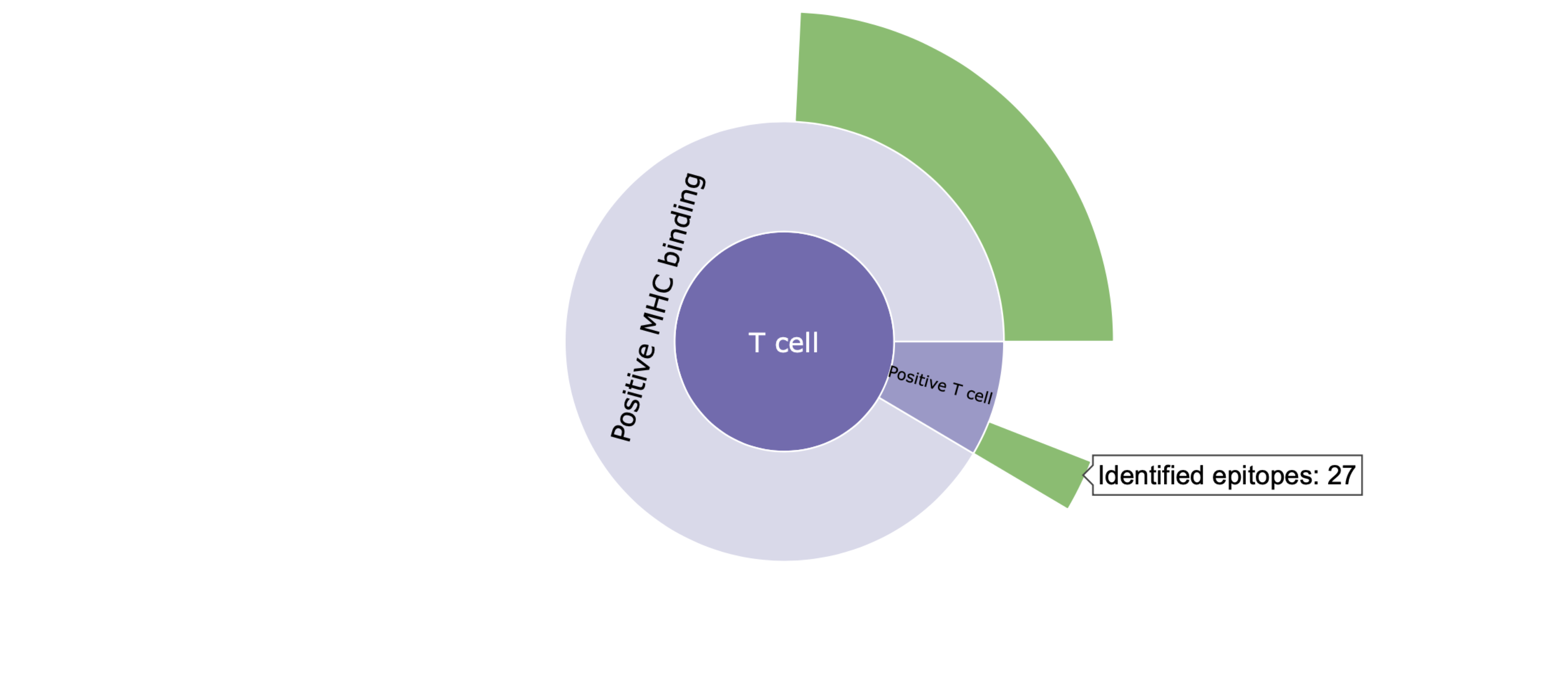
II. Clicking on a category level (e.g., T cell) in the sunburst plots focuses on the distribution of epitopes within that category. Clicking the same category again takes you back to the parent category.
Slider input labelled 'Minimum conservation of epitopes within SARS-CoV-2 sequences'
Definition of conservation: Conservation of an epitope within SARS-CoV-2 sequences represents the fraction of available SARS-CoV-2 sequences with the exact epitope sequence.
Identified B cell/T cell epitopes
Tables of epitopes
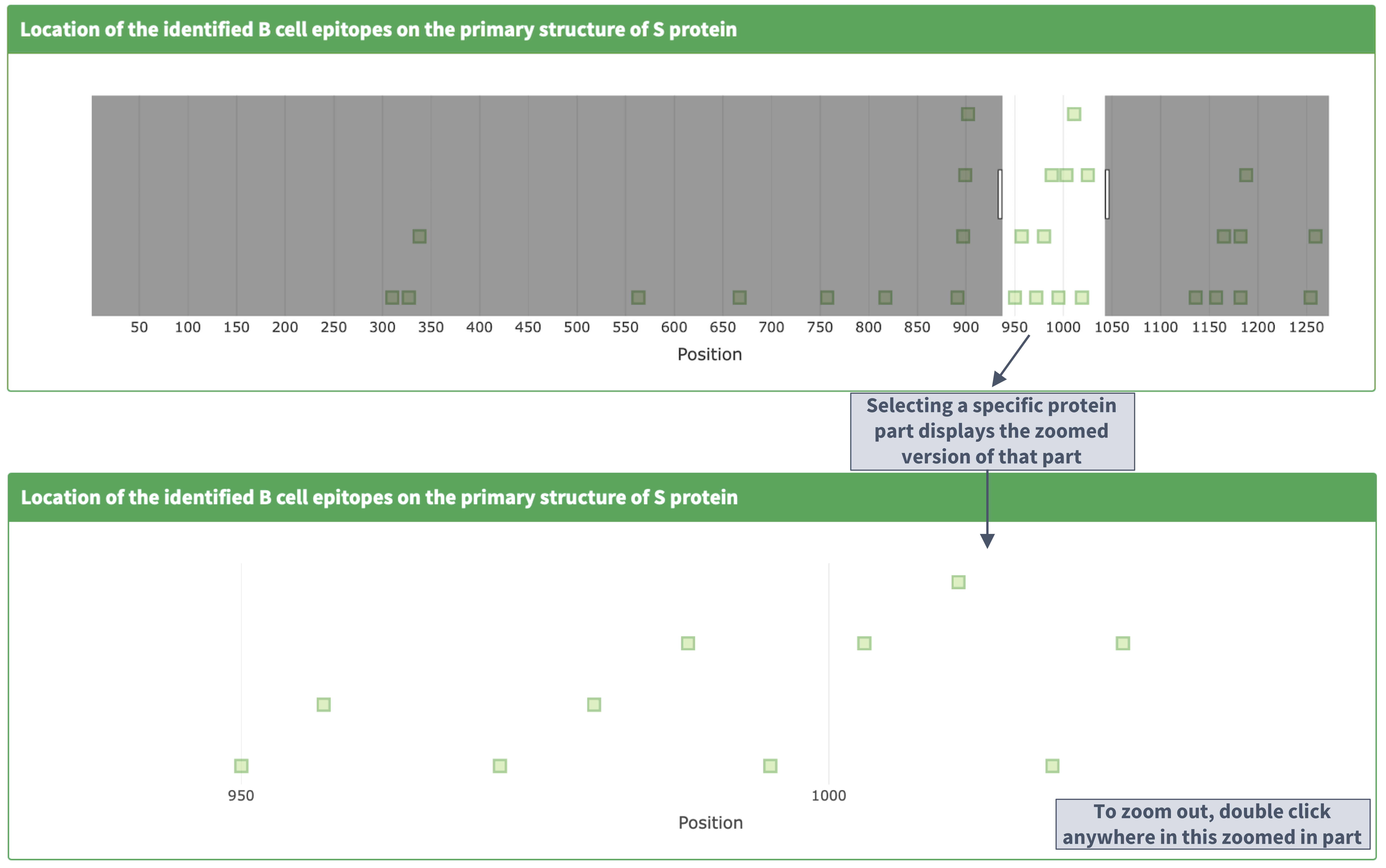
Description of the 'identified epitopes location on the primary structure' plot
I. Plot details
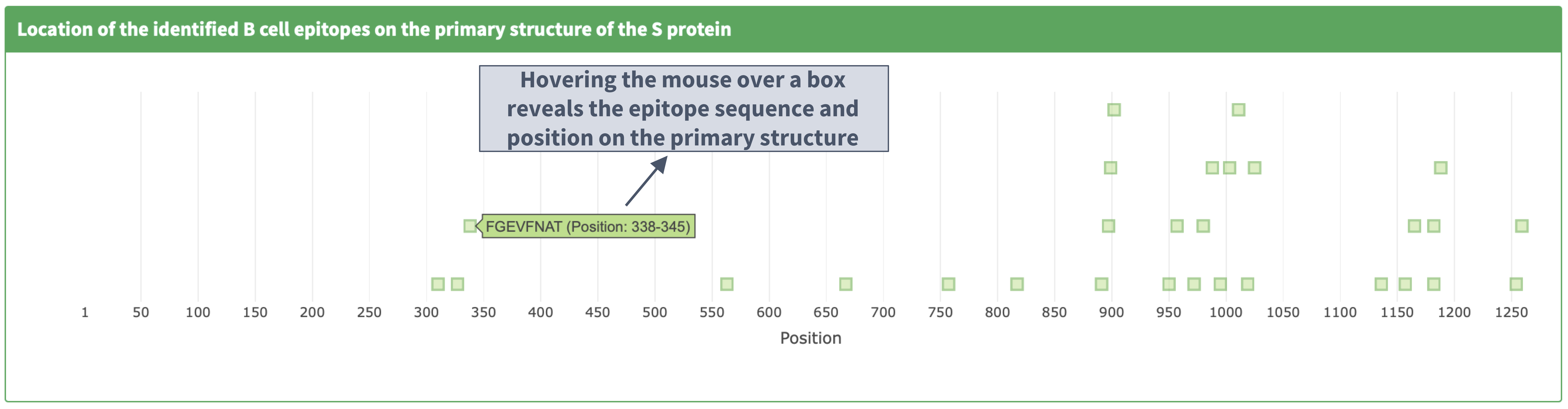
II. Zooming in and out on the primary structure
Population coverage analysis

Observed mutations in SARS-CoV-2 proteins
I. Plot details
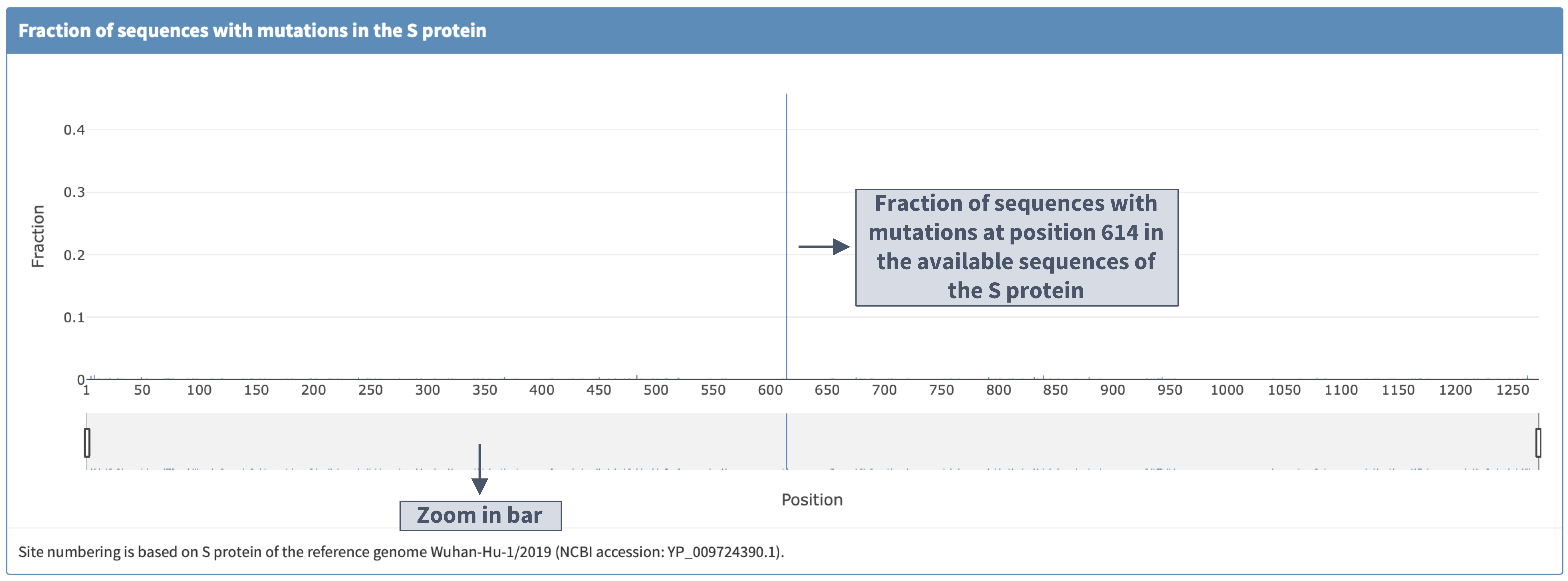
II. Zooming in and clicking the bar displays the mutations observed so far at a protein location
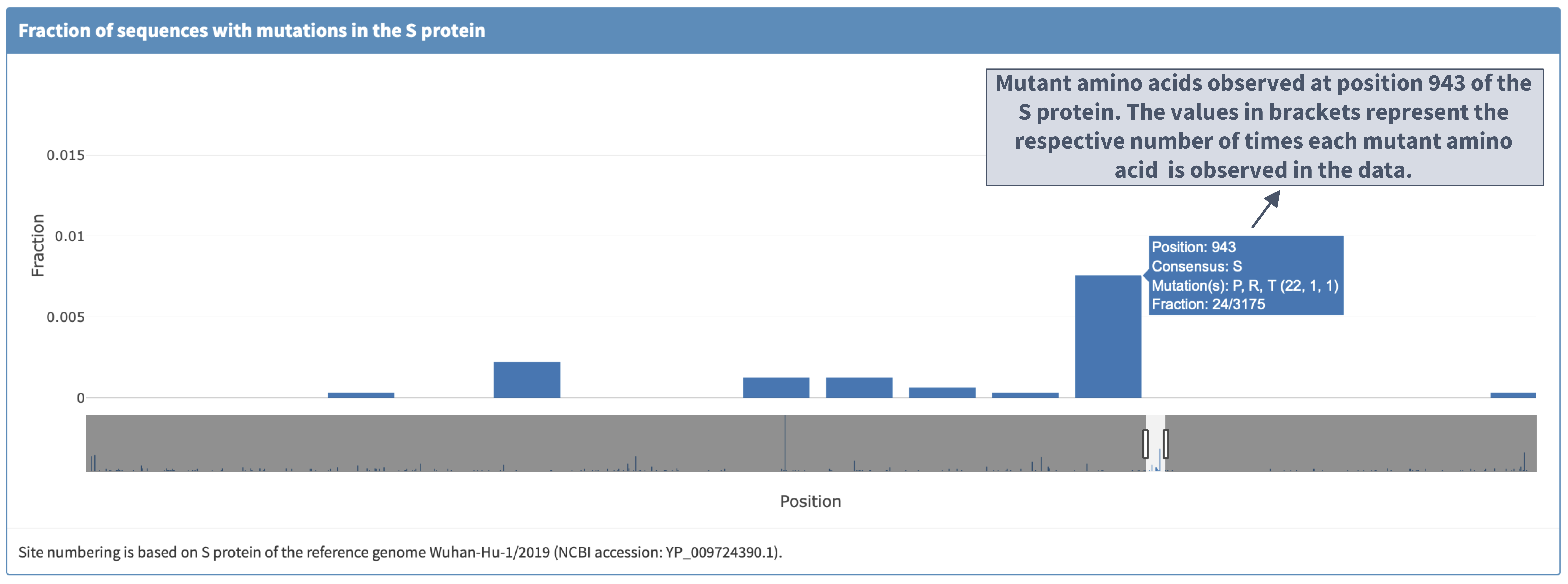
III. 'Reference sequence' plot

COVIDep development
COVIDep is developed and maintained by the Signal Processing & Computational Biology Lab ( SPCB ) at the Hong Kong University of Science and Technology (HKUST).
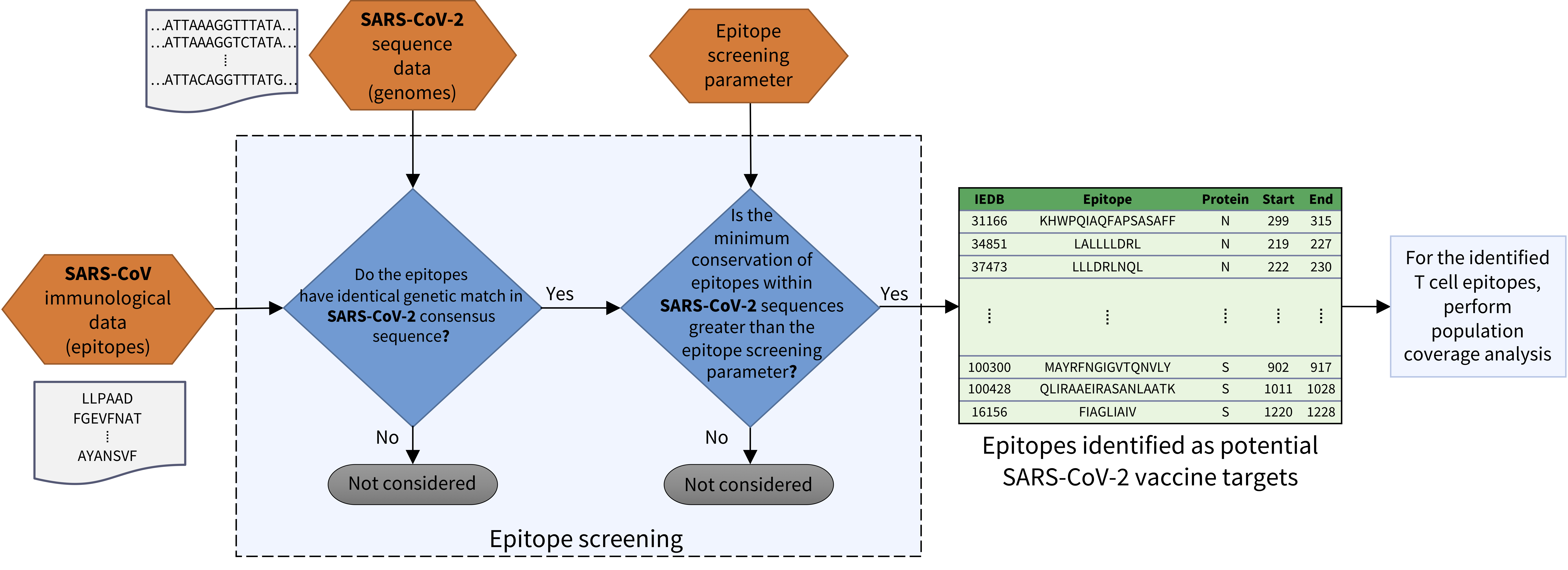
COVIDep aims to provide real-time potential vaccine targets for SARS-CoV-2. It screens the SARS-derived B cell and T cell epitopes (available at VIPR / IEDB ) and identifies those which are highly conserved within the available SARS-CoV-2 sequences (continuing to be deposited at GISAID ).
The raw genome sequences from 'Human' hosts are downloaded from GISAID and filtered to include only the full-length genomes. The sequences which are marked in the 'Comments' column on the GISAID website as having issues are excluded. All remaining genomic sequences are aligned and translated to yield protein sequences according to the reference genome of SARS-CoV-2 obtained from NCBI. The minimum conservation for selecting epitopes is determined by the global “epitope screening” parameter, which can be set using the slider in the menu bar. For the identified T cell epitopes, an estimated population coverage is computed using the population coverage tool provided at the IEDB Analysis Resource .
COVIDep is developed using the Shiny based web app development framework provided by RStudio. It is an open source web app and all the code scripts are available at the GitHub repository. The web app display is optimized for viewing at 100% screen resolution on Chrome for PC/laptop. For any questions, comments or suggestions, please feel free to contact us at COVIDep@ust.hk.
How to cite COVIDep
Ahmed, S.F., Quadeer, A.A. & McKay, M.R. COVIDep: a web-based platform for real-time reporting of vaccine target recommendations for SARS-CoV-2. Nature Protocols (2020). https://doi.org/10.1038/s41596-020-0358-9
Method
Related publications
Ahmed, S. F., Quadeer, A. A., & McKay, M. R. (2020). COVIDep platform for real-time reporting of vaccine target recommendations for SARS-CoV-2: Description and connections with COVID-19 immune responses and preclinical vaccine trials. bioRxiv, 2020.05.23.111385 https://www.biorxiv.org/content/10.1101/2020.05.23.111385v3
Ahmed, S. F., Quadeer, A. A., & McKay, M. R. (2020). Preliminary identification of potential vaccine targets for the COVID-19 coronavirus (SARS-CoV-2) based on SARS-CoV immunological studies. Viruses, 12(3), 254. Viruses. https://www.mdpi.com/1999-4915/12/3/254
Braun, J., Loyal, L., Frentsch, M., Wendisch, D., Georg, P., Kurth, F., Hippenstiel, S., et al. (2020). Presence of SARS-CoV-2 reactive T cells in COVID-19 patients and healthy donors. medRxiv, 2020.04.17.20061440. http://medrxiv.org/content/early/2020/04/22/2020.04.17.20061440
Gao, A., Chen, Z., Segal, F. P., Carrington, M., Streeck, H., Chakraborty, A. K., & Julg, B. (2020). Predicting the Immunogenicity of T cell epitopes: From HIV to SARS-CoV-2. bioRxiv, 2020.05.14.095885 https://www.biorxiv.org/content/10.1101/2020.05.14.095885v2
Chour, W., Xu, A. M., Ng, A. H. C., Choi, J., Xie, J., Yuan, D., Lee, J. K., et al. (2020). Shared antigen-specific CD8+ T cell responses against the SARS-COV-2 spike protein in HLA A*02:01 COVID-19 participants. medRxiv, 2020.05.04.20085779. http://medrxiv.org/content/early/2020/05/08/2020.05.04.20085779
Forcelloni, S., Benedetti, A., Dilucca, M., & Giansanti, A. (2020). Identification of conserved epitopes in SARS-CoV-2 spike and nucleocapsid protein. bioRxiv, 2020.05.14.095133. http://biorxiv.org/content/early/2020/05/15/2020.05.14.095133
Gao, Q., Bao, L., Mao, H., Wang, L., Xu, K., Yang, M., Li, Y., et al. (2020). Development of an inactivated vaccine candidate for SARS-CoV-2. Science, 21(1) https://science.sciencemag.org/content/early/2020/05/06/science.abc1932
Grifoni, A., Sidney, J., Zhang, Y., Scheuermann, R. H., Peters, B., & Sette, A. (2020). A sequence homology and bioinformatic approach can predict candidate targets for immune responses to SARS-CoV-2. Cell Host & Microbe, 27, 1–10. https://doi.org/10.1016/j.chom.2020.03.002
Grifoni, A., Weiskopf, D., Ramirez, S. I., Mateus, J., Jennifer, M., Moderbacher, C. R., Rawlings, S. A., et al. (2020). Targets of T cell responses to SARS-CoV-2 coronavirus in humans with COVID-19 disease and unexposed individuals. Cell. https://doi.org/10.1016/j.cell.2020.05.015
Ou, X., Liu, Y., Lei, X., Li, P., Mi, D., Ren, L., Guo, L., et al. (2020). Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nature Communications, 11(1), 1620. http://www.ncbi.nlm.nih.gov/pubmed/32221306
Pinto, D., Park, Y., Beltramello, M., Walls, A. C., Tortorici, M. A., Bianchi, S., Jaconi, S., et al. (2020). Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody. Nature. http://dx.doi.org/10.1038/s41586-020-2349-y
Poh, C. M., Carissimo, G., Bei, W., Amrun, S. N., Lee, C. Y.-P., Chee, R. S.-L., Yeo, N. K.-W., et al. (2020). Potent neutralizing antibodies in the sera of convalescent COVID-19 patients are directed against conserved linear epitopes on the SARS-CoV-2 spike protein. bioRxiv, 2020.03.30.015461. http://biorxiv.org/content/early/2020/03/31/2020.03.30.015461
Smith, T. R. F., Patel, A., Ramos, S., Elwood, D., Zhu, X., Yan, J., Gary, E. N., et al. (2020). Immunogenicity of a DNA vaccine candidate for COVID-19. Nature Communications, 11(1), 2601. http://dx.doi.org/10.1038/s41467-020-16505-0
Walls, A. C., Park, Y.-J., Tortorici, M. A., Wall, A., McGuire, A. T., & Veesler, D. (2020). Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell, 180, 1–12. http://www.ncbi.nlm.nih.gov/pubmed/32155444
Wang, C., Li, W., Drabek, D., Okba, N. M. A., van Haperen, R., Osterhaus, A. D. M. E., van Kuppeveld, F. J. M., et al. (2020). A human monoclonal antibody blocking SARS-CoV-2 infection. Nature Communications, 11(1), 2251. http://dx.doi.org/10.1038/s41467-020-16256-y
Wrapp, D., Wang, N., Corbett, K. S., Goldsmith, J. A., Hsieh, C.-L., Abiona, O., Graham, B. S., et al. (2020). Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science, 367(6483), 1260–1263. https://www.sciencemag.org/lookup/doi/10.1126/science.abb2507
Yin, D., Ling, S., Tian, X., Li, Y., Xu, Z., Jiang, H., Zhang, X., et al. (2020). A single dose SARS-CoV-2 simulating particle vaccine induces potent neutralizing activities. bioRxiv, 2020.05.14.093054. http://biorxiv.org/content/early/2020/05/15/2020.05.14.093054
Yu, J., Tostanoski, L. H., Peter, L., Mercado, N. B., McMahan, K., Mahrokhian, S. H., Nkolola, J. P., et al. (2020). DNA vaccine protection against SARS-CoV-2 in rhesus macaques. Science. https://www.sciencemag.org/lookup/doi/10.1126/science.abc6284
Zheng, M., & Song, L. (2020). Novel antibody epitopes dominate the antigenicity of spike glycoprotein in SARS-CoV-2 compared to SARS-CoV. Cellular & Molecular Immunology. http://dx.doi.org/10.1038/s41423-020-0385-z
Zheng, Z., Monteil, V. M., Maurer-Stroh, S., Yew, C. W., Leong, C., Mohd-Ismail, N. K., Arularasu, S. C., et al. (2020). Monoclonal antibodies for the S2 subunit of spike of SARS-CoV cross-react with the newly-emerged SARS-CoV-2. bioRxiv, 2020.03.06.980037. http://biorxiv.org/content/early/2020/03/07/2020.03.06.980037